GADMA with custom model (momi)
GADMA could be run with a detailed specified demographic model. Model should be provided as a file with a function model_func that describes demographic history by engine itself. In case of momi engine function should take values of parameters and return momi’s demographic model.
In this example we will take data and custom demographic model and infer parameters with usage of GADMA and momi engine.
Data
We will infer the demographic history of three classical modern human populations:
YRI - African population (Yorubans),
CEU - European population (Utah Residents (CEPH) with Northern and Western European Ancestry),
CHB - Asian population (Han Chinese in Beijing, China).
Data will be taken from VCF file. momi engine can read any type of SFS data including data for dadi and fastsimcoal2. However, opposite to dadi and moments momi engine requires full SFS data without any downsizing. That is why it is better to read the SFS data directly from VCF.
Our data includes SNP’s on 22 chromosome and was simulated with stdpopsim using msprime engine under the demographic history from Gutenkunst et al. 2009. Two individuals for each population are observed.
out_of_africa_chr22_sim.vcf- VCF file with chr22.out_of_africa_chr22_sim.popmap- file with (sample-population) correspondence.
Demographic model
Demographic model is located in demographic_model.py and is written according to `momi manual <https://momi2.readthedocs.io/en/latest/>`__.
It is important to note that units of parameters for momi engine are physical by default. Time for one generation should be equal to 1 and time should be in generations. If we want to mark that one parameter is in genetic units then we add _gen at the end of its name (e.g. nu_2F_gen).
Time parameters should be used (like usually in dadi) as time of epochs. This means that past events should occur in sum of several time parameters (as time of ancestral population split happened at moment T2+T3).
[1]:
%%bash
cat demographic_model.py
import momi
import numpy as np
def model_func(params):
"""
N_anc: Ancestral population size
N_1F: Size of ancestral population after growth and YRI population
N_2B: Bottleneck size of Eurasian population
nu_2F_gen: Final size of CEU population (in genetic units)
r2: Growth rate for CEU population
N_3F: Final size of CHB population
r3: Growth rate for CHB population
T1: Time of epoch between ancestral size growth and first split
T2: Time of second epoch between two splits
T3: Time of last epoch, time of last split
"""
N_Anc, N_1F, N_2B, nu_2F_gen, r2, N_3F, r3, T1, T2, T3 = params
model = momi.DemographicModel(N_e=1e5)
# momi2 uses backward in time models
# 1. Set leafs at time = 0
model.add_leaf("YRI", N=N_1F)
# we have nu_2F_gen is in genetic units so we translate it
model.add_leaf("CEU", N=nu_2F_gen * N_Anc, g=r2)
model.add_leaf("CHB", N=N_3F, g=r3)
# 2. Merge CHB to CEU (T3 time ago) and set size to bottleneck with g=0
model.move_lineages("CHB", "CEU", t=T3, N=N_1F, g=0)
# 3. Merge CEU to YRI (T3+T2 time ago)
model.move_lineages("CEU", "YRI", t=T3+T2)
# 4. Change size of ancestral (YRI) population to N_anc (T3+T2+T1 time ago)
model.set_size("YRI", N=N_Anc, g=0, t=T3+T2+T1)
return model
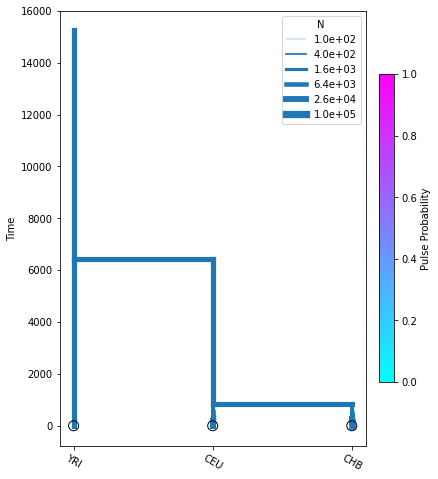
We could use some random parameters to plot this demographic model with momi:
[2]:
from demographic_model import model_func
import momi
rand_pars = [7300, 12300, 2100, 4, 0.004, 54090, 0.0055, 8800, 5600, 848]
model = model_func(rand_pars)
fig = momi.DemographyPlot(
model, ["YRI", "CEU", "CHB"],
figsize=(6,8)
)
Inference with GADMA
To run GADMA with custom model one should set parameters file:
[3]:
%%bash
cat params_file
# Set data first
Input file: out_of_africa_chr22_sim.vcf, out_of_africa_chr22_sim.popmap
# Output folder. It should be empty.
Output directory: gadma_result
# Set engine for simulations. We use momi.
Engine: momi
# For engine momi mutation rate and sequence length are required
Mutation rate: 1.29e-8
Sequence length: 51304566
# Now set our custom demographic model from file.
# There should be a function model_func in the file with a model.
Custom filename: demographic_model.py
# for short example
global_maxiter: 30
local_maxiter: 2
# How many repeats to run and how many processes to use.
Number of repeats: 2
Number of processes: 2
[4]:
%%bash
# As output directory should be empty we remove it if it exists
rm -rf gadma_result
Now we can run GADMA:
[5]:
%%bash
gadma -p params_file
Data reading
Number of populations: 3
Projections: [4, 4, 4]
Population labels: ('YRI', 'CEU', 'CHB')
Outgroup: True
--Successful data reading--
--Successful arguments parsing--
Parameters of launch are saved in output directory: /home/katenos/Workspace/popgen/temp/GADMA/examples/custom_model_momi/gadma_result/params_file
All output is saved in output directory: /home/katenos/Workspace/popgen/temp/GADMA/examples/custom_model_momi/gadma_result/GADMA.log
--Start pipeline--
Run launch number 1
Run launch number 2
[000:01:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244949.17 (N_Anc=192820.74841, N_1F=7223.22565, N_2B=346.23931, nu_2F_gen=0.23018, r2=6e-05, N_3F=2552.33854, r3=-3.98e-04, T1=495272.96232, T2=2424.316, T3=114.44749) mmmmmmm
Run 1 -247290.30 (N_Anc=8599.06242, N_1F=14879.82964, N_2B=17816.44499, nu_2F_gen=2.78733, r2=0.001, N_3F=2011.56609, r3=-1.56e-04, T1=805.41691, T2=5827.67398, T3=64.43284) mmmmmm
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:02:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244910.58 (N_Anc=192820.74841, N_1F=7223.22565, N_2B=664.04023, nu_2F_gen=0.24092, r2=0.00011, N_3F=1587.17085, r3=-3.98e-04, T1=753712.78309, T2=2424.316, T3=84.81883) mmmmmm
Run 1 -244974.98 (N_Anc=8096.63797, N_1F=5371.01532, N_2B=3719.21832, nu_2F_gen=0.48689, r2=0.00084, N_3F=440.22857, r3=-3.39e-05, T1=167.17264, T2=1748.29924, T3=10.3562) mmmmmmm
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:03:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244897.75 (N_Anc=308684.05537, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.42549, r2=0.00011, N_3F=2351.44351, r3=-6.54e-04, T1=1117566.07943, T2=2004.24654, T3=56.36652) mmmmmmmm
Run 1 -244974.98 (N_Anc=8096.63797, N_1F=5371.01532, N_2B=3719.21832, nu_2F_gen=0.48689, r2=0.00084, N_3F=440.22857, r3=-3.39e-05, T1=167.17264, T2=1748.29924, T3=10.3562) mmmmmmm
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:04:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244763.99 (N_Anc=49608.92298, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.07268, r2=0.00011, N_3F=2787.10375, r3=-7.05e-04, T1=360037.9623, T2=2424.316, T3=58.65638) c
Run 1 -244974.98 (N_Anc=8096.63797, N_1F=5371.01532, N_2B=3719.21832, nu_2F_gen=0.48689, r2=0.00084, N_3F=440.22857, r3=-3.39e-05, T1=167.17264, T2=1748.29924, T3=10.3562) mmmmmmm
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:05:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059)
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:06:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059)
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:07:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059)
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:08:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059)
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
[000:09:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059)
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
Finish genetic algorithm number 1
[000:10:00]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938)
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059) f
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
Finish genetic algorithm number 2
[000:10:10]
All best by log-likelihood models
Number log-likelihood Model
Run 2 -244627.21 (N_Anc=15004.59662, N_1F=7223.22565, N_2B=270.24291, nu_2F_gen=0.06908, r2=0.0009, N_3F=1820.32828, r3=-8.74e-04, T1=360037.9623, T2=2424.316, T3=36.21938) f
Run 1 -244902.38 (N_Anc=7320.66763, N_1F=8819.88308, N_2B=7340.6777, nu_2F_gen=0.1009, r2=0.001, N_3F=3287.38175, r3=-6.63e-05, T1=4398.86362, T2=2490.0807, T3=8.88059) f
Run 2 warning: failed to draw model due to the following exception: Model <class 'gadma.models.custom_demographic_model.CustomDemographicModel'> is not supported by demes engine.
The supported models are: [<class 'gadma.models.demographic_model.EpochDemographicModel'>, <class 'gadma.models.structure_demographic_model.StructureDemographicModel'>]..
You can find the Python code of the best model in the output directory.
--Finish pipeline--
You didn't specify theta at the beginning. If you want to change it and rescale parameters, please see the tutorial:
https://gadma.readthedocs.io/en/latest/user_manual/theta.html
Thank you for using GADMA!
In case of any questions or problems, please contact: ekaterina.e.noskova@gmail.com
UserWarning: Setting `Input file` is renamed in 2 version of GADMA to `Input data`. It is successfully read. (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/settings_storage.py:902)
UserWarning: Momi engine does not support Linear size function. It is removed from possible dynamics (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/settings_storage.py:1260)
UserWarning: Domain of DynamicVariable changed to ['Sud', 'Exp'] (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/settings_storage.py:617)
UserWarning: Momi engine need ancestral size as parameter. The option `Ancestral size as parameter` is set to `True` (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/settings_storage.py:1265)
UserWarning: Momi engine does not support continous migrations. The option `No migrations` is set to `True` (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/settings_storage.py:1271)
WARNING:momi.data.snps:No BED provided, will need to specify length manually with mutation rate
[W::vcf_parse] INFO 'AA' is not defined in the header, assuming Type=String
[W::vcf_parse] FILTER 'NOTPASS' is not defined in the header
UserWarning: Setting no_migrations (True) will be ignored as custom model from file is chosen. (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/arg_parser.py:293)
UserWarning: Setting split_fractions (True) will be ignored as custom model from file is chosen. (/home/katenos/.local/lib/python3.6/site-packages/gadma/cli/arg_parser.py:293)
It is just a start of optimization as numer of iterations is limited to 30 but we can see that size of ancestral population is similar to 7300 that was used for simulation. We can evaluate value of log-likelihood on simulated values:
[6]:
# load data
ind2pop = {'tsk_0': 'YRI', 'tsk_1': 'YRI', 'tsk_2': 'CEU', 'tsk_3': 'CEU', 'tsk_4': 'CHB', 'tsk_5': 'CHB'}
data = momi.SnpAlleleCounts.read_vcf('out_of_africa_chr22_sim.vcf', ind2pop=ind2pop).extract_sfs(n_blocks=100)
data = data.subset_populations(['YRI', 'CEU', 'CHB'])
# evaluate log likelihood
model.set_data(data, length=51304566)
ll_model = model.log_likelihood()
print(f'Value of log-likelihood on values used in simulation: {ll_model}')
No BED provided, will need to specify length manually with mutation rate
Value of log-likelihood on values used in simulation: -249931.26661084342
[ ]: